Prior and (noisy) posterior predictions#
Show the difference between noisy and noiseless predictions by plotting. Now we generate 1\(D\) toy data \((x_i\in\mathbb R, y_i\in\mathbb R)\) and also perform a “real GP fit” by optimizing the \(\ell\) hyperparameter. However we fix \(\sigma_n^2\) to certain values to showcase the different noise cases.
Below \(\ma\Sigma\equiv\cov(\ve f_*)\) is the covariance matrix from [RW06] eq. 2.24 with \(\ma K = K(X,X)\), \(\ma K'=K(X_*, X)\) and \(\ma K''=K(X_*, X_*)\), so
Imports and helpers
Generate 1\(D\) toy data
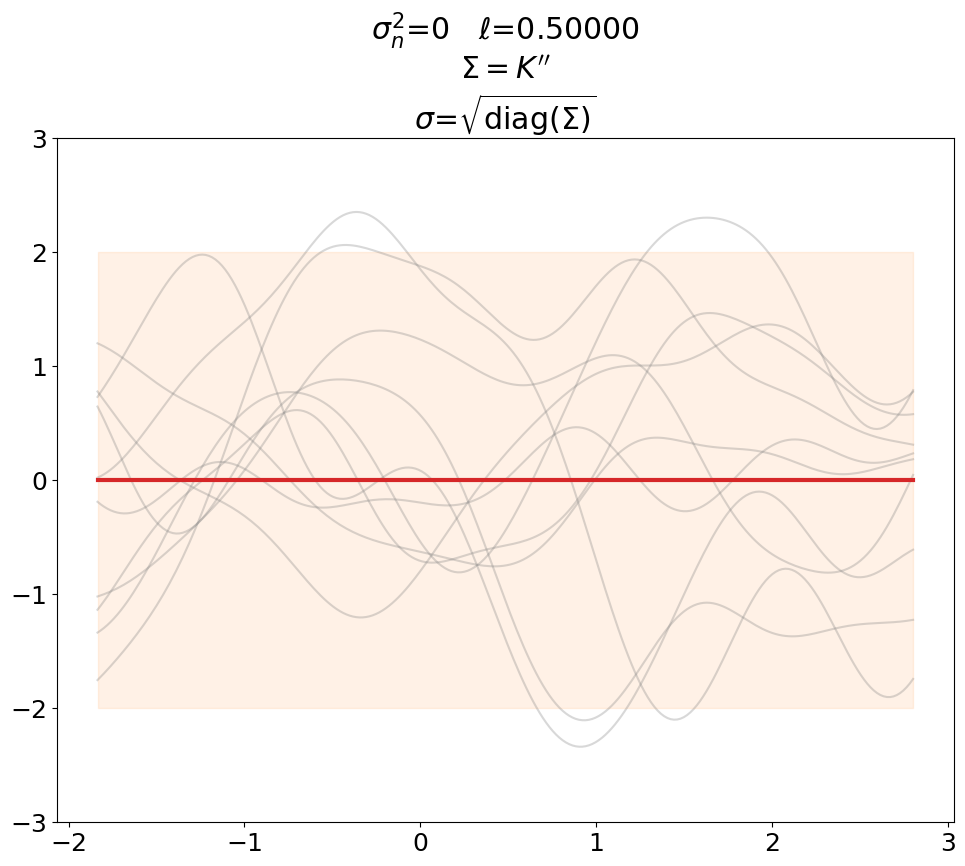
Prior, noiseless#
First we plot the prior, without noise (predict_noiseless).
This is the standard textbook case. We set \(\ell\) to some constant of our liking since its value is not defined in the absence of training data.
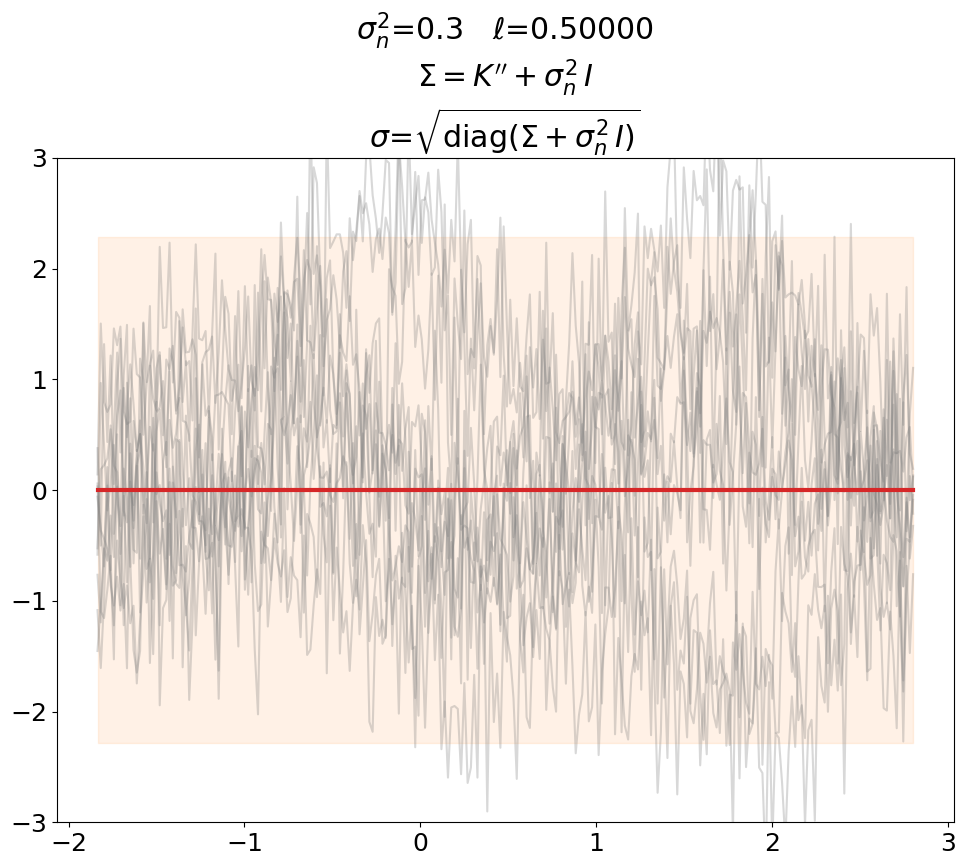
Prior, noisy#
Even though not super useful, we can certainly generate noisy prior samples
in the predict setting when using \(\ma K'' + \sigma_n^2\,\ma I\) as prior
covariance.
This is also shown in [DLvdW20] in fig. 4.
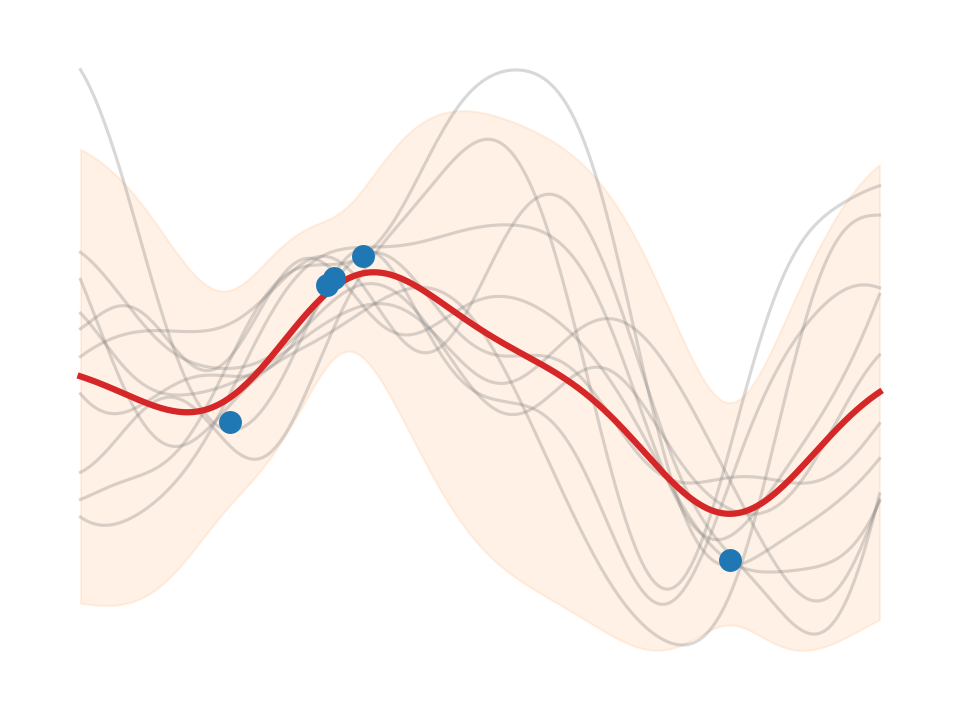
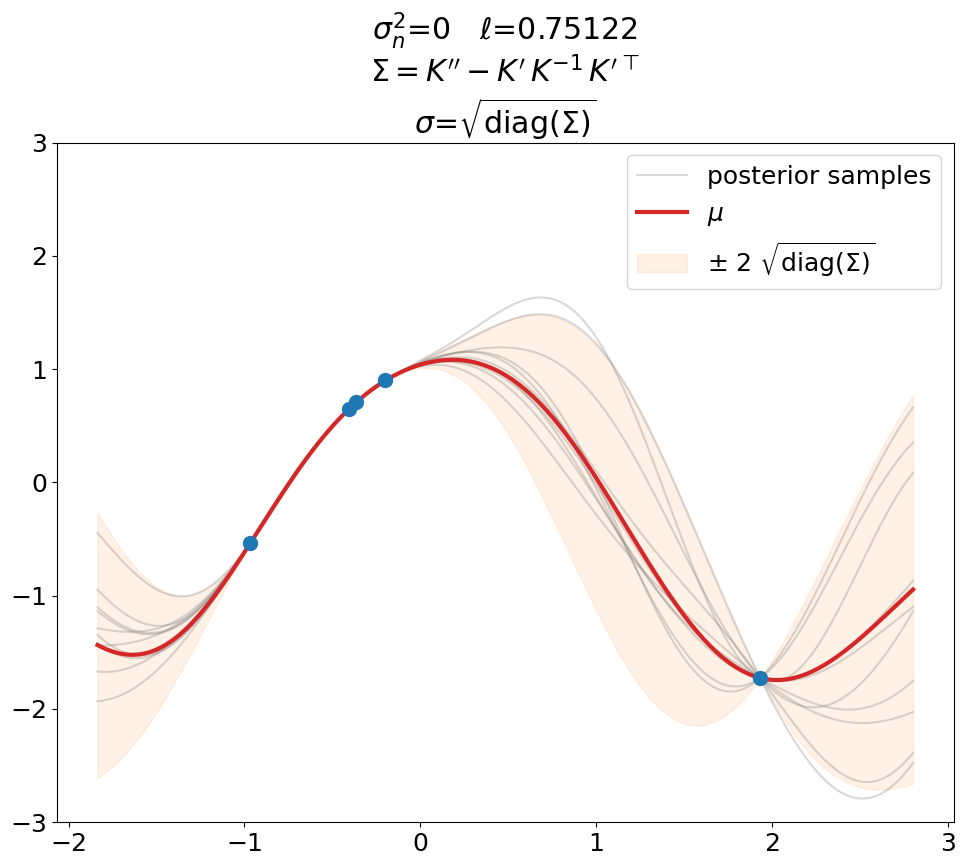
Posterior, noiseless, interpolation#
Now the posterior.
For that, we do an \(\ell\) optimization using 1\(D\) toy data and sklearn
with fixed \(\sigma_n^2\):
interpolation (\(\sigma_n = 0\))
regression (\(\sigma_n > 0\))
Interpolation (\(\sigma_n^2=0\)).
This is a plot you will see in most text books. Notice that \(\ell\) is not 0.5 as above but has been optimized (maximal LML).
Posterior, noiseless, regression#
Regression (\(\sigma_n^2>0\)), predict_noiseless.
You will see a plot like this also in text books. For instance
[Mur23] has this in fig.
18.7 (book version 2022-10-16). If you inspect the code that generates it
(which is open source, thanks!), you find that they use tinygp in the
predict_noiseless setting.
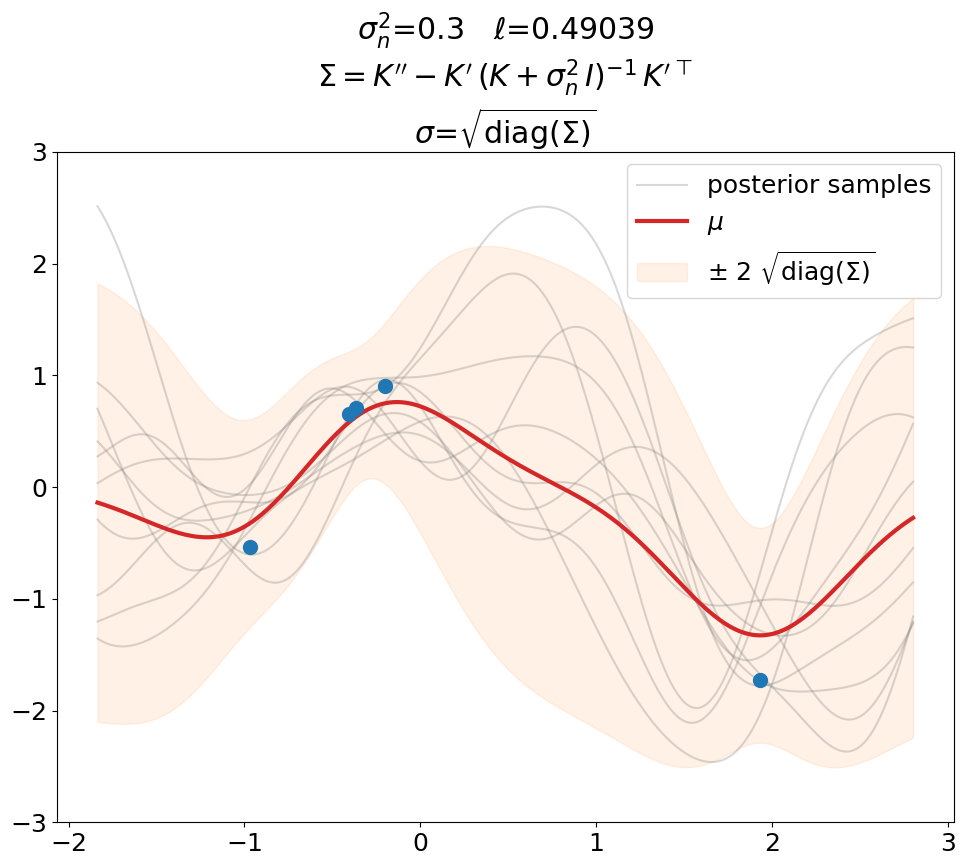
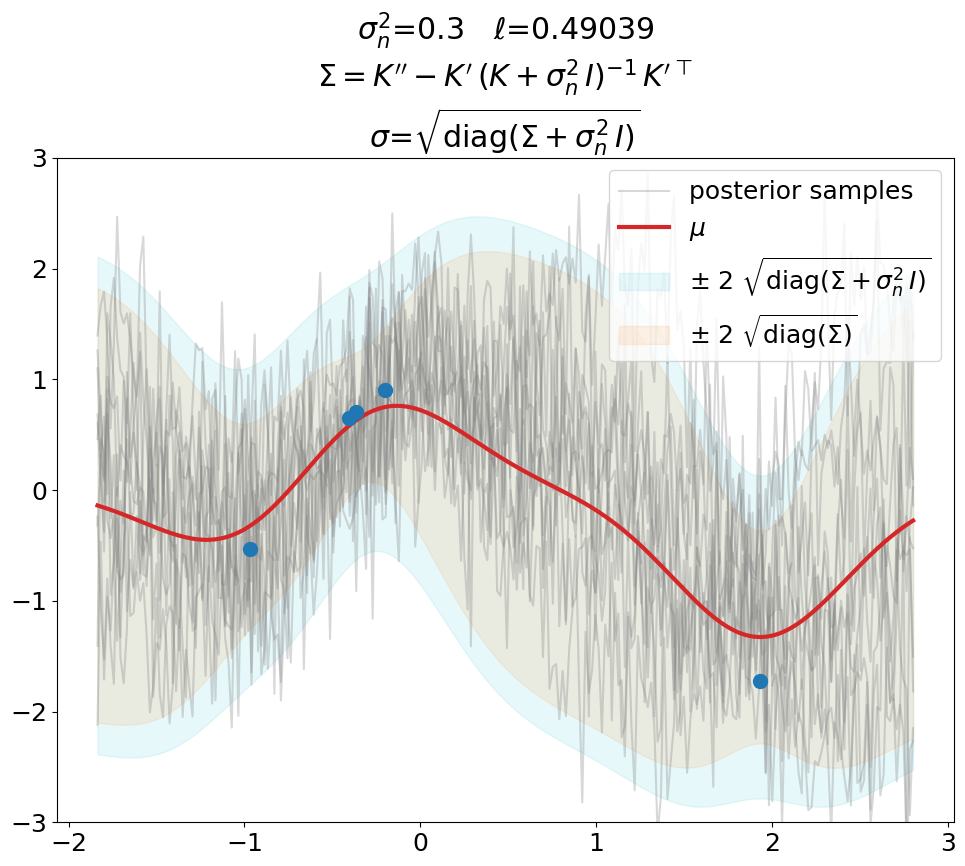
Posterior, noisy, regression#
Regression (\(\sigma_n^2>0\)), predict.
This is the same as above in terms of optimized \(\ell\), \(\sigma_n^2\) value (fixed here, usually also optimized), fit weights \(\ve\alpha\) and thus predictions \(\ve\mu\).
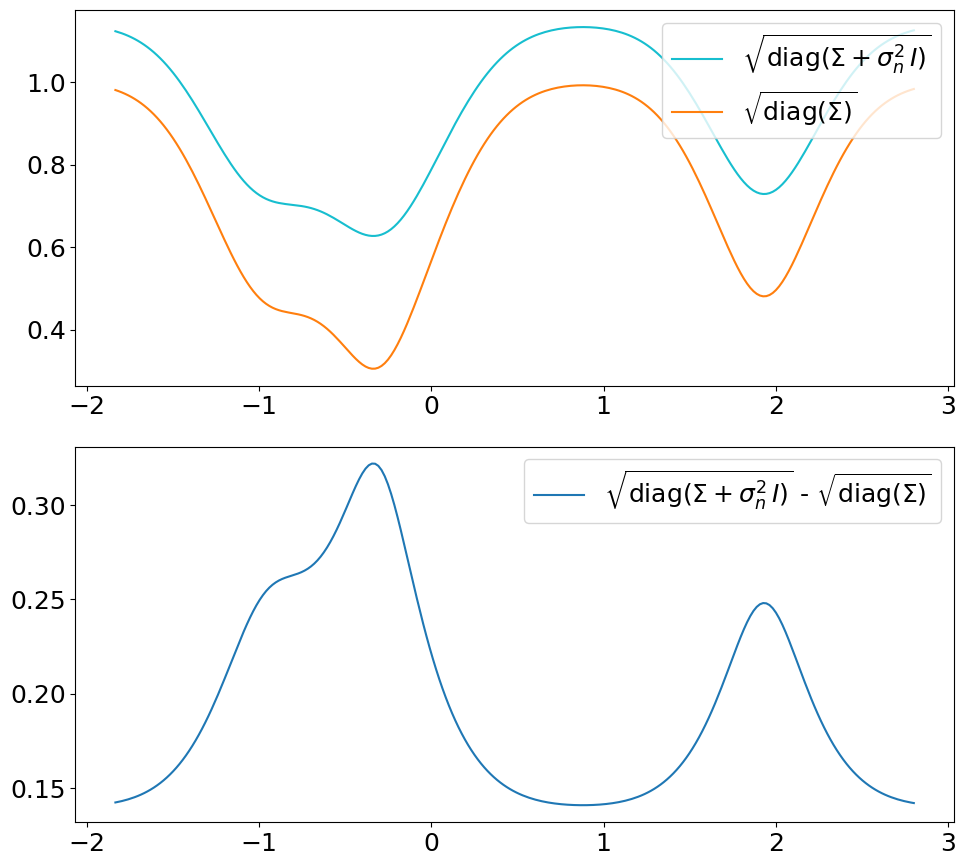
The only difference is that we now use the covariance matrix \(\ma\Sigma + \sigma_n^2\,\ma I\) instead of \(\ma\Sigma\) to generate samples from the posterior. By that we get (1) noisy samples and (2) a larger \(\sigma\). That’s what is typically not shown in text books (at least not the ones we checked). Noisy samples (only for the prior, but still) are shown for instance in [DLvdW20] in fig. 4.
The difference in \(\sigma\) between predict vs. predict_noiseless is not
constant even though the constant \(\sigma_n^2\) is added to the diagonal
because of the \(\sqrt{\cdot}\) in